ABSTRACT
Human hematopoietic/stem progenitor CD34+ cells (HSPC) are the core components of ex vivo lentiviral gene therapy (GT) for the treatment of rare monogenic diseases. Understanding the composition and dynamics of this population upon ex-vivo therapeutic modification is critical in determining the potency of GT drug products (DP). To overcome the limitations presented by the use of standard surface markers upon ex vivo gene transfer, we combined immunophenotyping, fluorescence-activated cell sorting (FACS), and in vitro manipulation with single-cell RNA-Sequencing (scRNA-Seq), capturing a picture of unprecedented clarity on CD34+ subsets dynamics after culture. Using the 10X Chromium System, we first generated a comprehensive baseline map of CD34+ cell states (~80,000 single cells) from the 3 main sources of human HSPC: bone marrow (BM), mobilized peripheral blood (G-CSF or G-CSF+Plerixafor mobilization, MPB) and umbilical cord blood (CB), unveiling key differences in HSPC composition depending on the cells’ origin. We then focused on G-CSF+Plerixafor MPB, and studied at single-cell resolution the dynamic changes of clinical-grade genetically engineered CD34+ cells, as well as FACS-sorted and individually cultured HSPC subsets. With this approach, we were able to measure a substantial amount of cells maintaining an hematopoietic stem cell (HSC)-like transcriptome in the DP manufactured with our proprietary system. We could also evaluate for the first time the extent of myelo-megakaryo-erythroid differentiation occurring in cytokine-stimulated primitive and committed progenitors. Notably, our comprehensive dataset (total of 24,736 and 56,391 single cells at baseline and end of culture, respectively) represents a unique resource that we are interrogating for the identification of novel markers able to capture the true DP composition at the end of culture. Three novel HSC-specific candidate markers are currently undergoing wetlab validation. Overall, our analytical platform provides an advanced suite for the characterization of CD34+ cells states with broad application spanning manufacturing, and pre-clinical and clinical stages.
Human hematopoietic/stem progenitor CD34+ cells (HSPC) are the core components of ex vivo lentiviral gene therapy (GT) for the treatment of rare monogenic diseases. Understanding the composition and dynamics of this population upon ex-vivo therapeutic modification is critical in determining the potency of GT drug products (DP). To overcome the limitations presented by the use of standard surface markers upon ex vivo gene transfer, we combined immunophenotyping, fluorescence-activated cell sorting (FACS), and in vitro manipulation with single-cell RNA-Sequencing (scRNA-Seq), capturing a picture of unprecedented clarity on CD34+ subsets dynamics after culture. Using the 10X Chromium System, we first generated a comprehensive baseline map of CD34+ cell states (~80,000 single cells) from the 3 main sources of human HSPC: bone marrow (BM), mobilized peripheral blood (G-CSF or G-CSF+Plerixafor mobilization, MPB) and umbilical cord blood (CB), unveiling key differences in HSPC composition depending on the cells’ origin. We then focused on G-CSF+Plerixafor MPB, and studied at single-cell resolution the dynamic changes of clinical-grade genetically engineered CD34+ cells, as well as FACS-sorted and individually cultured HSPC subsets. With this approach, we were able to measure a substantial amount of cells maintaining an hematopoietic stem cell (HSC)-like transcriptome in the DP manufactured with our proprietary system. We could also evaluate for the first time the extent of myelo-megakaryo-erythroid differentiation occurring in cytokine-stimulated primitive and committed progenitors. Notably, our comprehensive dataset (total of 24,736 and 56,391 single cells at baseline and end of culture, respectively) represents a unique resource that we are interrogating for the identification of novel markers able to capture the true DP composition at the end of culture. Three novel HSC-specific candidate markers are currently undergoing wetlab validation. Overall, our analytical platform provides an advanced suite for the characterization of CD34+ cells states with broad application spanning manufacturing, and pre-clinical and clinical stages.
COMMENTARY
INTRODUCTION
Hematopoietic stem cell GT for rare monogenic diseases is based on the autologous transplantation of CD34+ cells transduced ex vivo with a lentiviral vector (LVV) carrying a functional copy of the defective gene [1]. The resulting CD34+ DP, after re-infusion and engraftment in patients, will generate a progeny of gene corrected cells expressing the therapeutic protein, which will exert a potentially lifelong correction of the genetic defect. CD34+ cells are composed by functionally heterogeneous progenitor subsets, traditionally defined by the expression of surface markers such as CD38, CD90, CD45RA, CD7, CD10, CD135, CD49f [2, 3]. We previously showed how these markers do not allow a clear cut identification of the diversity of cell states within the CD34+ compartment [4]; moreover, because of the instability of such markers in culture [5, 6] immunophenotyping does not offer a reliable assay to measure the HSPC content within the CD34+ DP. To overcome these limitations, we leveraged scRNA-Seq technologies as the most advanced methods available for resolving cell populations’ heterogeneity, gaining new insight on CD34+ cells composition at steady state and their dynamics upon ex vivo manipulation.
Hematopoietic stem cell GT for rare monogenic diseases is based on the autologous transplantation of CD34+ cells transduced ex vivo with a lentiviral vector (LVV) carrying a functional copy of the defective gene [1]. The resulting CD34+ DP, after re-infusion and engraftment in patients, will generate a progeny of gene corrected cells expressing the therapeutic protein, which will exert a potentially lifelong correction of the genetic defect. CD34+ cells are composed by functionally heterogeneous progenitor subsets, traditionally defined by the expression of surface markers such as CD38, CD90, CD45RA, CD7, CD10, CD135, CD49f [2, 3]. We previously showed how these markers do not allow a clear cut identification of the diversity of cell states within the CD34+ compartment [4]; moreover, because of the instability of such markers in culture [5, 6] immunophenotyping does not offer a reliable assay to measure the HSPC content within the CD34+ DP. To overcome these limitations, we leveraged scRNA-Seq technologies as the most advanced methods available for resolving cell populations’ heterogeneity, gaining new insight on CD34+ cells composition at steady state and their dynamics upon ex vivo manipulation.
MATERIALS & METHODS
Cryopreserved, healthy donor-derived CD34+ cells were purchased from AllCells or Hemacare. For FACS analysis, CD34+ cells were thawed according to manufacturer instructions and stained for CD34, CD38, CD90, CD45RA, CD7, CD10, CD135 and lineage markers as previously described [4]. Immunophenotyping was performed on a Symphony A3 Analyzer (BD Biosciences); cell sorting was performed on a Sony MA900 instrument; flow cytometry data were analyzed with FlowJo 10.7.2 software. Research grade, bulk CD34+ or FACS-sorted HSPC subsets were cultured ex vivo in the presence of rhSCF (300ng/mL), rhTPO (100ng/mL), rhFLT3-L (300ng/mL), rhIL3 (60ng/mL) (StemCell Technologies). Clinical grade CD34+ DP was manufactured from fresh GCSF+Plerixafor mobilized leukopack (AllCells), through AVROBIO’s proprietary plato® gene therapy platform; a cryopreserved aliquot of the apheresis product and final DP were thawed and processed for immunophenotyping and single cell analysis. For scRNA-seq, cells were processed with Chromium Next GEM Single Cell 3ʹ Reagent Kits v3.1 (10X Genomics), according to manufacturer instructions; single-cell libraries were sequenced with Illumina NextSeq 500/550 High Output Kit v2.5 (150 Cycles) or NovaSeq S2 platforms. Sequence processing was performed using the CellRanger suite (10X Genomics) and downstream analyses were conducted via Seurat and Monocle R packages.
Cryopreserved, healthy donor-derived CD34+ cells were purchased from AllCells or Hemacare. For FACS analysis, CD34+ cells were thawed according to manufacturer instructions and stained for CD34, CD38, CD90, CD45RA, CD7, CD10, CD135 and lineage markers as previously described [4]. Immunophenotyping was performed on a Symphony A3 Analyzer (BD Biosciences); cell sorting was performed on a Sony MA900 instrument; flow cytometry data were analyzed with FlowJo 10.7.2 software. Research grade, bulk CD34+ or FACS-sorted HSPC subsets were cultured ex vivo in the presence of rhSCF (300ng/mL), rhTPO (100ng/mL), rhFLT3-L (300ng/mL), rhIL3 (60ng/mL) (StemCell Technologies). Clinical grade CD34+ DP was manufactured from fresh GCSF+Plerixafor mobilized leukopack (AllCells), through AVROBIO’s proprietary plato® gene therapy platform; a cryopreserved aliquot of the apheresis product and final DP were thawed and processed for immunophenotyping and single cell analysis. For scRNA-seq, cells were processed with Chromium Next GEM Single Cell 3ʹ Reagent Kits v3.1 (10X Genomics), according to manufacturer instructions; single-cell libraries were sequenced with Illumina NextSeq 500/550 High Output Kit v2.5 (150 Cycles) or NovaSeq S2 platforms. Sequence processing was performed using the CellRanger suite (10X Genomics) and downstream analyses were conducted via Seurat and Monocle R packages.
FIGURE 1
scRNA profile and immuno-phenotype of bulk CD34+ cells at steady state and upon ex vivo engineering
scRNA profile and immuno-phenotype of bulk CD34+ cells at steady state and upon ex vivo engineering
A) Global CD34+ reference map
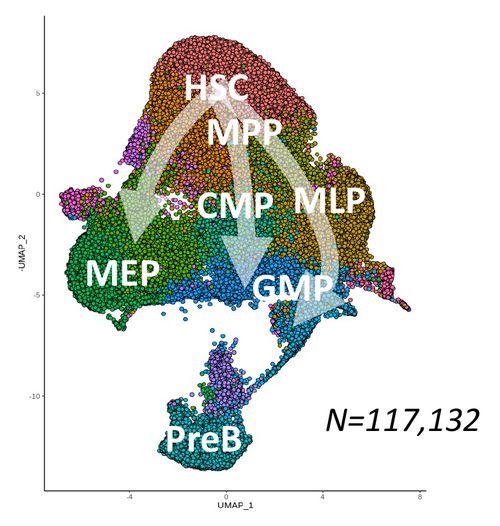
HSC: Hematopoietic Stem Cells
MPP: Multipotent Progenitors
MLP: Multilymphoid Progenitor
ETP: Early Thymic Progenitor
PreB-NK: Precursor of B and NK cells
CMP: Common Myeloid Progenitor
GMP: Granulo-Monocyte Progenitor
MEP: Megakaryo-Erythroid Progenitor
LIN+: Lineage positive cells
MPP: Multipotent Progenitors
MLP: Multilymphoid Progenitor
ETP: Early Thymic Progenitor
PreB-NK: Precursor of B and NK cells
CMP: Common Myeloid Progenitor
GMP: Granulo-Monocyte Progenitor
MEP: Megakaryo-Erythroid Progenitor
LIN+: Lineage positive cells
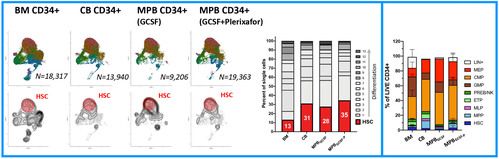
B) CD34+ cell sources
C) CD34+ Gene Therapy DP
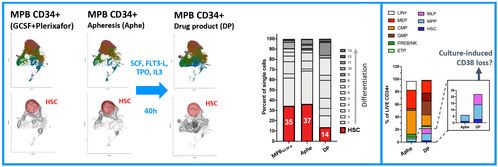
FIGURE 2
scRNA profile and immuno-phenotype of FACS-sorted and individually cultured hematopoietic subsets
scRNA profile and immuno-phenotype of FACS-sorted and individually cultured hematopoietic subsets
A) Sorting strategy
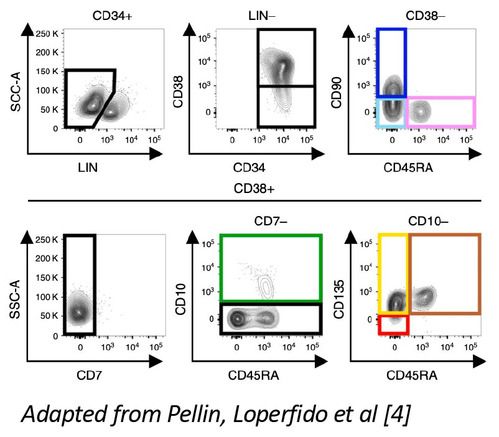
B) Immunophenotype of individual HSPC subsets before and after transduction
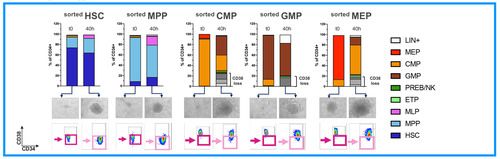
C) scRNA-Seq of individual HSPC subsets before and after transduction
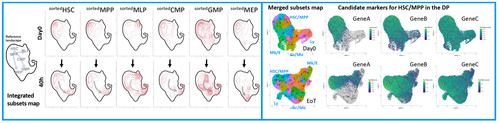
RESULTS & DISCUSSION
We firstly generated a global reference map of single CD34+ cells (Fig.1A) obtained from different sources: BM (the natural site of hematopoiesis in the adult), CB (a source reflective of more primitive hematopoiesis) and mobilized peripheral blood (the most advanced and clinically relevant source of CD34+ cells for GT). We calculated the fraction of HSC-related transcriptomes in each sample, observing that GCSF+Plerixafor MPB were the ones with higher content of HSC-like cells (Fig.1B). This is in line with the superior engraftment reported for this CD34 cell source [7, 8] as compared to BM and further supports its use as a source of drug product in GT clinical trials. Immunophenotypic analysis on these steady-state samples overall confirmed source-specific differences in HSCs and committed progenitors composition.
We next focused on the changes occurring upon therapeutic gene transfer and performed scRNAseq and FACS-immunophenotyping on GCSF+Plerixafor MPB before and at the end of a proprietary run of genetic engineering. The analysis was not only conducted on the bulk CD34+ population but also on 7 individually sorted progenitor cells kept in culture separately in the same conditions. At the end of transduction, we observed an increase of events in the committed progenitors compartment of the single-cell map (Fig.1B), which is an expected effect of the cytokines used during transduction; despite that, we still measured a substantial amount of cells maintaining an HSC-related transcriptome in the final DP. Conversely, FACS-immunophenotyping suggested an enrichment in phenotypically primitive progenitors at the end of transduction: our next experiment, performed on sorted hematopoietic progenitors, shed more light on this discrepancy.
We indeed observed that committed HSPCs, originally sorted as CD38+ subsets (CMP, GMP, MEP) start losing CD38 expression already at 40h of culture, thus creating a confounding factor in the assessment of their relative proportion with the primitive CD38- subsets (HSC, MPP, MLP) in the DP (Fig.2A). On the other end, scRNA profiling of these samples allowed to map the differentiation trajectories occurring in each subpopulation, uncovering the extent of myelo, megakaryo and erythroid differentiation occurring in individual primitive and committed progenitors (Fig.2B). Lastly, unsupervised clustering of sorted HSPCs single-cell transcriptomes, allowed the identification of 7 novel signatures through which we could interrogate the dynamic changes of each subset looking for transcriptional divergences/similarities between the baseline and the end of culture cell states. Through this approach we have discovered novel candidate markers whose expression is specifically retained/activated by HSCs after culture. We are currently testing their use to estimate the content of long-term HSCs contained in the DP, a previously unavailable and much needed potency readout for ex vivo GT (Fig.2C)
We firstly generated a global reference map of single CD34+ cells (Fig.1A) obtained from different sources: BM (the natural site of hematopoiesis in the adult), CB (a source reflective of more primitive hematopoiesis) and mobilized peripheral blood (the most advanced and clinically relevant source of CD34+ cells for GT). We calculated the fraction of HSC-related transcriptomes in each sample, observing that GCSF+Plerixafor MPB were the ones with higher content of HSC-like cells (Fig.1B). This is in line with the superior engraftment reported for this CD34 cell source [7, 8] as compared to BM and further supports its use as a source of drug product in GT clinical trials. Immunophenotypic analysis on these steady-state samples overall confirmed source-specific differences in HSCs and committed progenitors composition.
We next focused on the changes occurring upon therapeutic gene transfer and performed scRNAseq and FACS-immunophenotyping on GCSF+Plerixafor MPB before and at the end of a proprietary run of genetic engineering. The analysis was not only conducted on the bulk CD34+ population but also on 7 individually sorted progenitor cells kept in culture separately in the same conditions. At the end of transduction, we observed an increase of events in the committed progenitors compartment of the single-cell map (Fig.1B), which is an expected effect of the cytokines used during transduction; despite that, we still measured a substantial amount of cells maintaining an HSC-related transcriptome in the final DP. Conversely, FACS-immunophenotyping suggested an enrichment in phenotypically primitive progenitors at the end of transduction: our next experiment, performed on sorted hematopoietic progenitors, shed more light on this discrepancy.
We indeed observed that committed HSPCs, originally sorted as CD38+ subsets (CMP, GMP, MEP) start losing CD38 expression already at 40h of culture, thus creating a confounding factor in the assessment of their relative proportion with the primitive CD38- subsets (HSC, MPP, MLP) in the DP (Fig.2A). On the other end, scRNA profiling of these samples allowed to map the differentiation trajectories occurring in each subpopulation, uncovering the extent of myelo, megakaryo and erythroid differentiation occurring in individual primitive and committed progenitors (Fig.2B). Lastly, unsupervised clustering of sorted HSPCs single-cell transcriptomes, allowed the identification of 7 novel signatures through which we could interrogate the dynamic changes of each subset looking for transcriptional divergences/similarities between the baseline and the end of culture cell states. Through this approach we have discovered novel candidate markers whose expression is specifically retained/activated by HSCs after culture. We are currently testing their use to estimate the content of long-term HSCs contained in the DP, a previously unavailable and much needed potency readout for ex vivo GT (Fig.2C)
CONCLUSION
By high-throughput sc-RNAseq of steady-state CD34+ cells we provided a comprehensive global reference of baseline CD34+ transcriptional states and uncovered source-specific differences in HSPCs composition. We then explored the changes in transcriptional state of CD34+ cells upon gene transfer. scRNAseq of clinical-grade GT CD34+ DP and individually cultured HSPC subsets allowed overcoming the limitations of FACS-immunophenotyping for assessing the composition of cultured cells, unlocking the discovery and validation of novel markers specifically expressed by subsets of interest in the DP. Overall, our analytical platform provides the basis for unravelling and comparing the impact of multiple conditions of cell cultures and gene modification on the CD34+ DP. To our knowledge this constitutes the most advanced suite for the comprehensive characterization of CD34+ cells states with broad application spanning manufacturing, pre-clinical and clinical stages.
By high-throughput sc-RNAseq of steady-state CD34+ cells we provided a comprehensive global reference of baseline CD34+ transcriptional states and uncovered source-specific differences in HSPCs composition. We then explored the changes in transcriptional state of CD34+ cells upon gene transfer. scRNAseq of clinical-grade GT CD34+ DP and individually cultured HSPC subsets allowed overcoming the limitations of FACS-immunophenotyping for assessing the composition of cultured cells, unlocking the discovery and validation of novel markers specifically expressed by subsets of interest in the DP. Overall, our analytical platform provides the basis for unravelling and comparing the impact of multiple conditions of cell cultures and gene modification on the CD34+ DP. To our knowledge this constitutes the most advanced suite for the comprehensive characterization of CD34+ cells states with broad application spanning manufacturing, pre-clinical and clinical stages.
REFERENCES
- Ferrari G, Thrasher AJ, Aiuti A. Gene therapy using haematopoietic stem and progenitor cells. Nat Rev Genet. 2021 Apr;22(4):216-234. doi: 10.1038/s41576-020-00298-5. Epub 2020 Dec 10. PMID: 33303992.
- Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012 Feb 3;10(2):120-36. doi: 10.1016/j.stem.2012.01.006. PMID: 22305562.
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011 Jul 8;333(6039):218-21. doi: 10.1126/science.1201219. PMID: 21737740.
- Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, Biffi A, Klein AM, Biasco L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. 2019 Jun 3;10(1):2395. doi: 10.1038/s41467-019-10291-0. PMID: 31160568; PMCID: PMC6546699.
- Spangrude GJ, Brooks DM, Tumas DB. Long-term repopulation of irradiated mice with limiting numbers of purified hematopoietic stem cells: in vivo expansion of stem cell phenotype but not function. Blood. 1995 Feb 15;85(4):1006-16. PMID: 7849289.
- Danet GH, Lee HW, Luongo JL, Simon MC, Bonnet DA. Dissociation between stem cell phenotype and NOD/SCID repopulating activity in human peripheral blood CD34(+) cells after ex vivo expansion. Exp Hematol. 2001 Dec;29(12):1465-73. doi: 10.1016/s0301-472x(01)00750-0. PMID: 11750106.
- Hassan HT, Zeller W, Stockschläder M, Krüger W, Hoffknecht MM, Zander AR. Comparison between bone marrow and G-CSF-mobilized peripheral blood allografts undergoing clinical scale CD34+ cell selection. Stem Cells. 1996 Jul;14(4):419-29. doi: 10.1002/stem.140419. PMID: 8843543.
- Miller PH, Nakamichi N, Knapp DJHF, Rabu G, Schultz KR, Jones DM, Couban S, Eaves CJ. Quantitation of Human Cells that Produce Neutrophils and Platelets in Vivo Obtained from Normal Donors Treated with Granulocyte Colony-Stimulating Factor and/or Plerixafor. Biol Blood Marrow Transplant. 2016 Nov;22(11):1945-1952. doi: 10.1016/j.bbmt.2016.07.023. Epub 2016 Aug 2. PMID: 27496214.
STATEMENTS
All authors are employees of AVROBIO, Inc.
This research was funded by AVROBIO, Inc.
This data in this poster was presented at ASH 2020. Published with permission from the Copyright owner.
All authors are employees of AVROBIO, Inc.
This research was funded by AVROBIO, Inc.
This data in this poster was presented at ASH 2020. Published with permission from the Copyright owner.
